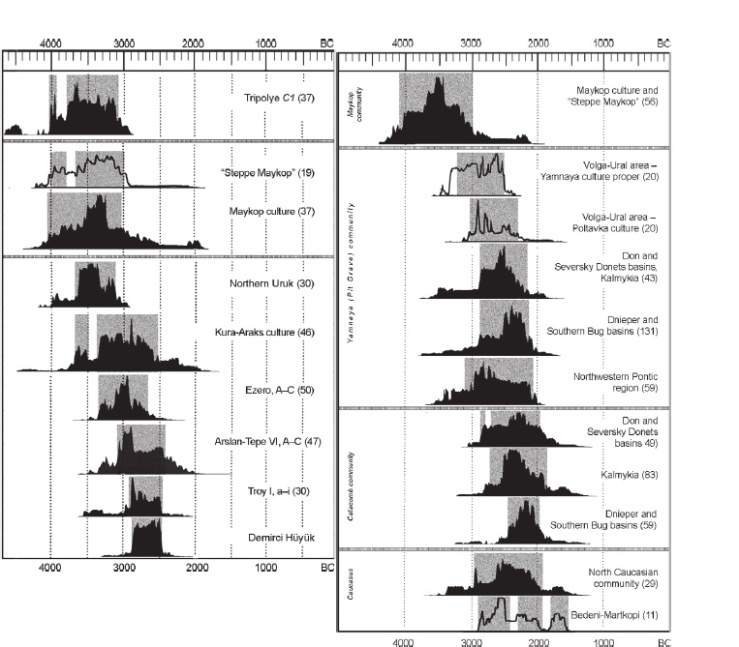
Thanks to Wang et al (2018), we got our first look at a group referred to as “Steppe Maykop”. Chernykh (2008) does briefly describe this group in his work, “Formation of the Eurasian “Steppe Belt” of stockbreeding cultures: viewed through the prism of archaeometallurgy and radiocarbon dating”. He goes on to explain that this is a kurgan using culture outside of the Maykop proper zone, north of the Kuban and Terek Basins, between the Sea of Azov and Caspian Sea (Chernykh, 2008). These steppe complexes contained Maykop cultural items (mostly pottery) (Chernykh, 2008). The image below, taken from Chernykh (2008), illustrates the location of Steppe Maykop.
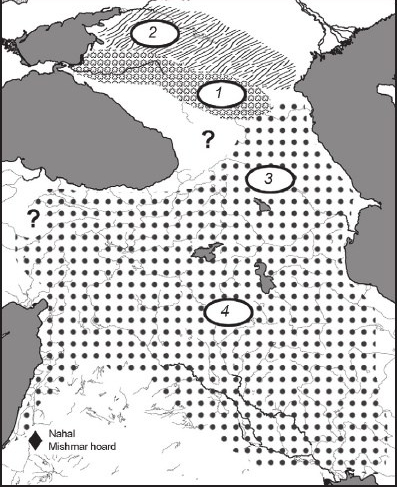
Fig. 4.
Circumpontic Metallurgical Province at the early formation stage (proto-CMP).
1 – Maykop culture proper;
2 – sites of the so-called Maykop type (“Steppe Maykop”);
3 – Kura-Araks culture;
4 – late (northern) sites of the Uruk community
DNA results of Steppe Maykop
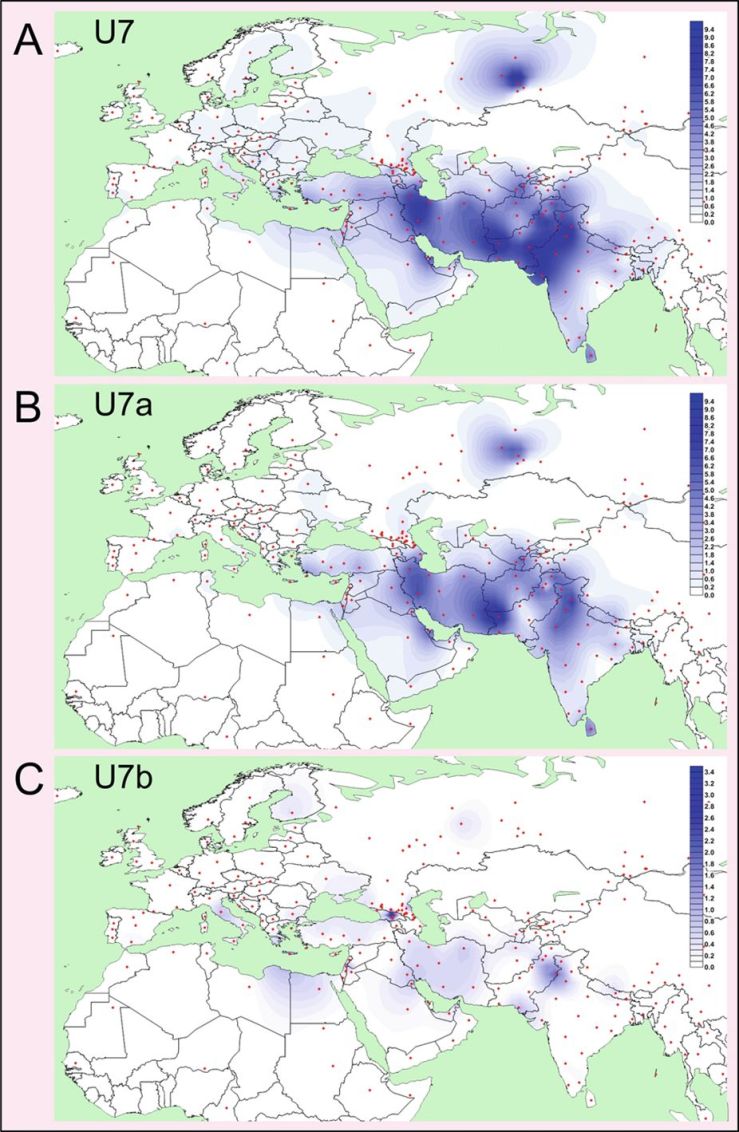
In the supplementary information of Wang et al (2018), the Steppe Maykop samples showed T2e, H2a1, and U7b. The one Y-DNA resulted in Q1a2. These are some interesting results, especially U7b and Q1a2.
U7b is likely rooted in West Asia between 5-10 KYA (Sahakyan et al, 2017), as it is mostly found in West Asia and Southeast Europe. It almost completely absent from Central Asia, where U7 and U7a are more typically found (Sahakyan et al, 2017). Ancient remains from Central Asia are also giving the same story, with U7 and U7a being the only ones found (Narasimhan et al, 2018). Also interesting is the hot spot of U7b in Georgia (Sahakyan et al, 2017), but this is based on modern populations. These Steppe Maykop samples are our first ancient samples with U7b. With their connection to Maykop, that may be the likely source of this haplogroup.
Q1a2 is a common haplogroup of Northern Asia and the Americas, showing that relationship to Native Americans that was discussed in Wang et al (2018). Today, it is fairly common among Turkic speakers of Siberia and Northern Central Asia, along with groups in the Americas.
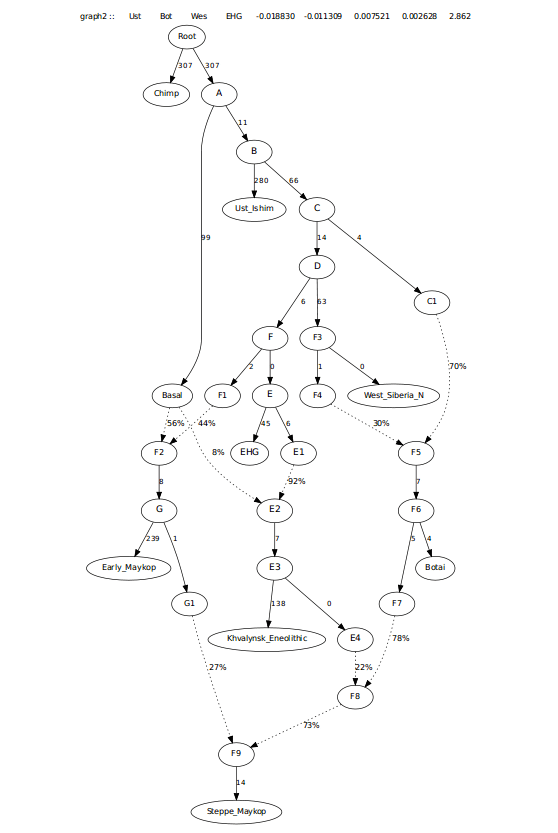
Modeling with qpGraph
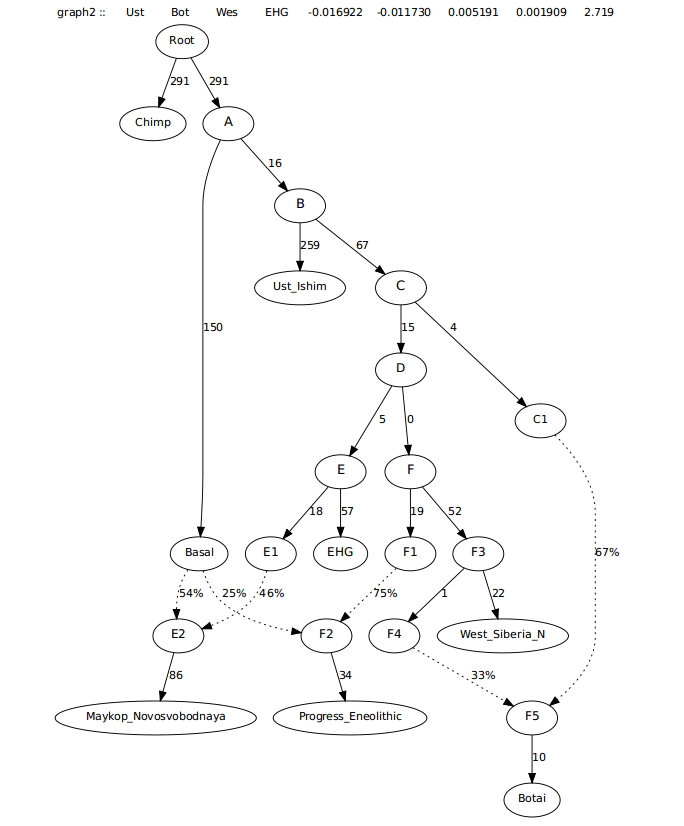
I started first by building a tree that included Chimp, Ust_Ishim, Botai, West_Siberia_N, Maykop_Novosvobodnaya, and Progress_Eneolithic. These samples come from Narasimhan et al (2018), Jeong et al (2018), and Wang et al (2018).
The next thing I did was add Steppe_Maykop to the column designating labels, but did not manually add them to any tree, to find the most significant scores for the first placement. The worst f-stat was Botai, Steppe_Maykop; West_Siberia_N Steppe_Maykop (Z>-49).
To determine if this stat was driven more by Botai or West_Siberia_N, I looked at the Z-scores where only the two were used (Botai, Steppe_Maykop; Botai, Steppe_Maykop and West_Siberia_N, Steppe_Maykop; West_Siberia_N, Steppe_Maykop).
The highest Z-score between the two was -42 between Botai and Steppe_Maykop, versus -30 for West_Siberia_N.
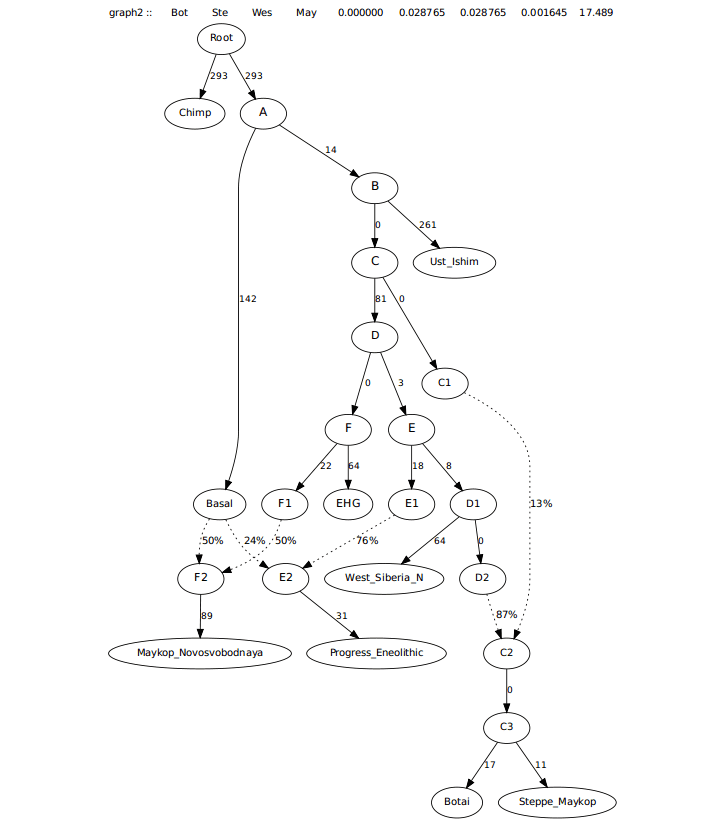
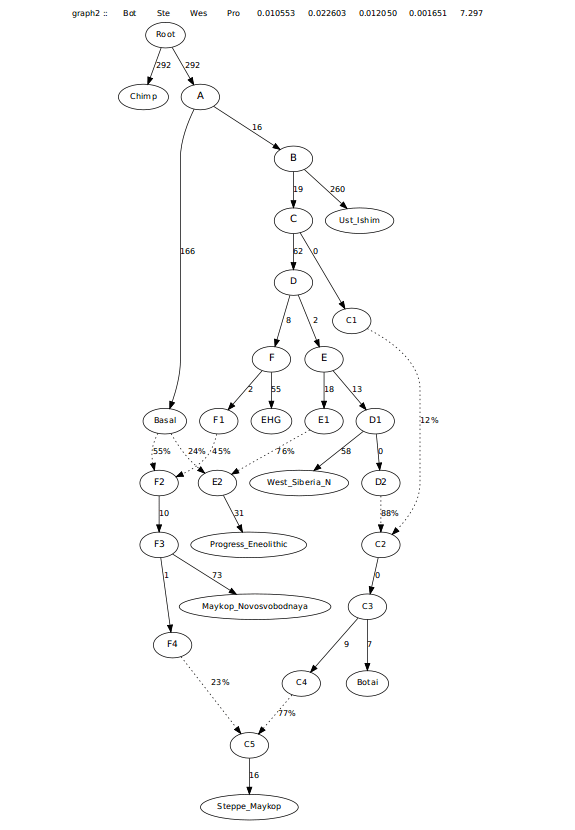
Trying to model this admixture as coming from just West_Siberia_N and not Botai left many 0 drift edges and a worst f-stat approaching 4, likely seeking admixture from Botai and more from Maykop, making it look less feasible. See first graph below. Second one below is the one where Steppe Maykop first branches off of Botai.
This graph left a worst f-stat asking for an edge from Progress_Eneolithic to Steppe_Maykop.
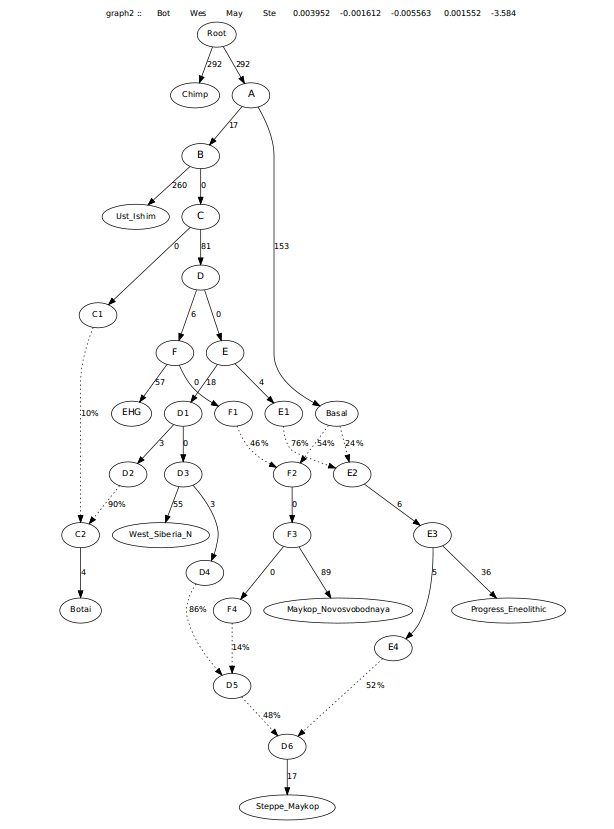
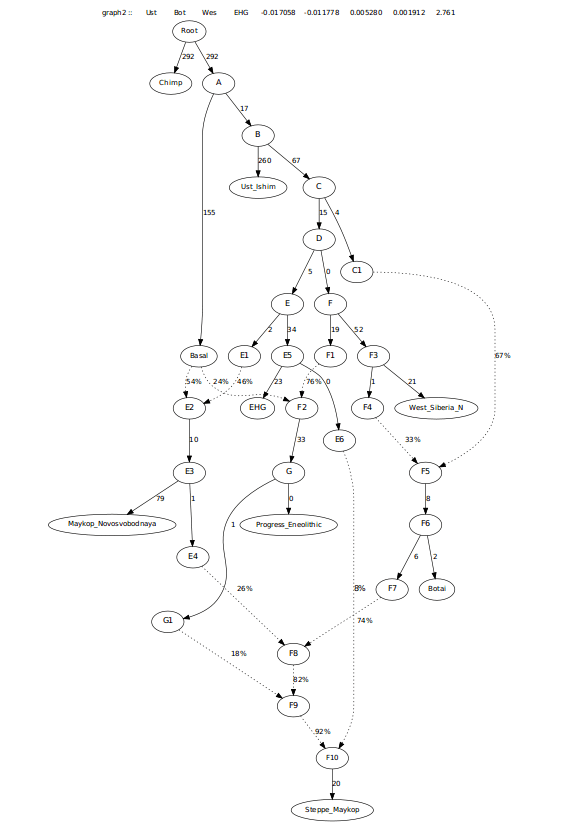
This last graph did have the Steppe_Maykop requiring more Maykop admixture than Progress_Eneolithic, more in-line with the f3-ratio stats (see below). However, I wanted to look at this another way as another way of looking at this created worst Z-scores asking for admixture from EHG, possibly pointing to this group have steppe ancestry that is more like Khvalynsk. Resulting in the tree below.
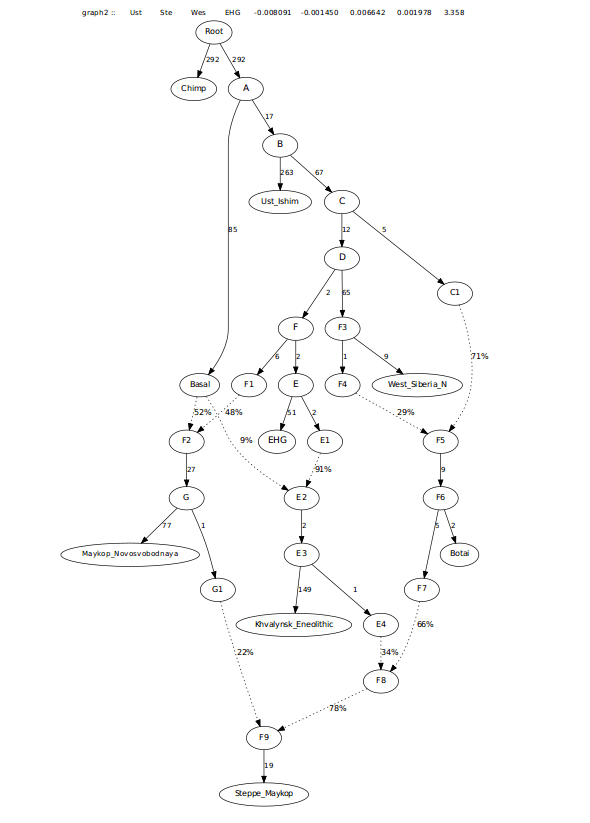
I also looked at this admixture potentially coming from Early_Maykop, as the f-3 score for both Maykop groups was similar. This also worked well.
This last one was more in-line with f3-ratio outputs and also resolved the need for extra EHG when removing the 0 drift edge in there.
f3-ratio
I decided to look at f3-ratio as well, to look for confirmation of these results. The results showed significant scores where the two sources were like Botai and Early_Maykop/Maykop_Novosvobodnaya. So, there is some good confirmation here that the models line up.
| Source 1 |
Source2 |
Target |
f_3 |
std. Error |
Z |
SNPs |
| Botai |
E_Maykop |
St_Maykop |
-0.008599 |
0.002107 |
-4.081 |
68545 |
| Botai |
Maykop_N |
St_Maykop |
-0.008444 |
0.001574 |
-5.364 |
126581 |
| Botai |
Progress |
St_Maykop |
-0.003914 |
0.001447 |
-2.705 |
141243 |
| Botai |
Khvalynsk |
St_Maykop |
0.005247 |
0.001706 |
3.075 |
106850 |
The f3-ratio using West_Siberia_N as the source 1 pop did produce more significant scores than Botai, but this could be due to the fact that Siberia_N is rather intermediary to Botai and Progress, as well as Khvalynsk.
Discussion
When looking back at all the data, combining the uni-parental markers, along with the statistical output, it seems that a population from around the Urals did move into the region north of Maykop proper, and acquired their cultural material (Chernykh, 2008), along with their genetic input. Since Maykop and Steppe Maykop both have the same carbon dates, it could be that Early Maykop is the better source, but there is limited samples here across that time-frame to really come to that conclusion. Progress and Khvalynsk, or something in-between make a good fit as the steppe ancestry in these ssamples. All-in-all it seems very possible for this to be the story of the genesis of Steppe Maykop, from a source more related to Native Americans, along with more typical West Asian ancestry.
Sources
Chernyk (2008) Formation of the Eurasian “Steppe Belt” of stock-breeding cultures. https://www.academia.edu/22557016/FORMATION_OF_THE_EURASIAN_STEPPE_BELT_OF_STOCKBREEDING_CULTURES.BY_E.N._Chernykh
Jeong et al (2018) Characterizing the genetic history of admixture across inner Eurasia. https://www.nature.com/articles/srep46044
Narasimhan et al (2018) The genetic formation of South and Central Asia. https://www.biorxiv.org/content/10.1101/292581v1
Sahakyan et al (2017) Origin and spread of human mitochondrial DNA haplogroup U7. https://www.nature.com/articles/srep46044
Wang et al (2018) The genetic history of the Greater Caucasus. https://www.biorxiv.org/content/10.1101/322347v1.supplementary-material